What sort of genetic outcome would be seen for each of the two theories when mtDNA is analyzed in present-day human populations? For multiregionalism, we would expect to see a very ancient origin or coalescence date for all extant mtDNA haplogroups, correlating more or less with the first migration from Africa by H. erectus. We would also expect to see deep-rooted genetic differences between populations which would reflect their antiquity and evolution from African, Asian, and European H. erectus populations.
With the Out of Africa or replacement model, the coalescence of mtDNA haplotypes would be much more recent, corresponding to the more recent origin of H. sapiens. We would expect to see less genetic diversity between populations of modern humans. We would also expect that the population which has the most intra-population genetic diversity would be the oldest, and hence the original population from which modern humans originated and migrated into the rest of the Old World. These two sets of predictions are eminently testable with genetic data.
MtDNA Evidence Supports Recent Evolution of H. sapiens
The first study to use mtDNA to investigate human evolution was published in 1987 by Rebecca Cann, Mark Stoneking, and the late Allan Wilson. They analyzed mtDNA from 147 individuals from a number of populations and constructed a phylogenetic tree. The root of the tree was placed in Africa - in other words, the ancestral mtDNA to all others was located in Africa, and by applying the molecular clock concept the ancestral mtDNAwas dated to c. 200 000 ya (within confidence limits of 140 000-290 000 ya). This ancestral mtDNA was dubbed Mitochondrial Eve by the media and some misunderstood this to mean that just one woman lived in Africa. This is certainly not the case - it means that other ancestral mtDNAs of other women did not leave female descendants to contribute their mtDNA to present-day humans. This initial study was criticized on a number of grounds - the phylogenetic methods used to analyze the data (several different trees could be constructed from the same data, with the root not always in Africa), the origin of the African mtDNA samples (many of these actually were obtained from Afro-Americans, so may not have been truly representative of African mtDNA haplotypes), and even the choice of mtDNA as the genetic locus of investigation.
Since this ground-breaking publication, many more genetic studies on human evolution have been carried out, using mtDNA, Y chromosome DNA and a wide variety of nuclear genes. Many of these studies show that the highest intrapopulation genetic diversity is found in Africa. High genetic diversity is found in both older populations (the older the population, the more DNA mutations become fixed) and expanding populations (the larger the population, the more DNA mutations become fixed). Some knowledge of Late Pleistocene human populations in the Old World is necessary in order to determine the correct interpretation. Estimates have been made of the size of populations required for each of the two theories. For example, using numbers based on hunter-gatherer groups, it has been estimated that the world population at around 150 000 ya was between 125 000 and 1 000 000. However, the effective population size consists of the actual number of individuals in a population that are reproducing, and is usually estimated to be one-third of the census population, that is, 42 000333 000 individuals throughout the Old World. The question is whether this would have been enough for the sort of gene flow between populations in Africa, Asia, and Europe envisaged by multiregionalists. If we consider the Out of Africa scenario, this would entail a smaller number of individuals drawn from a larger population, forming what is called a bottleneck (or founder event). Analysis of the overall low genetic diversity of modern humans has enabled an estimate to be made of the effective population size in this bottleneck - just 10000. This is almost a mystical number in human evolutionary genetics - it crops up time and again with different statistical analyses of genetic data - and it means that H. sapiens lived on a knife-edge of extinction for much of its early existence in Africa. Therefore, we can say that the high genetic diversity seen within African populations points to Africa as the place where modern humans evolved, and indicates the age, not the size, of the population.
Other Genetic Studies Support the Out of Africa Hypothesis
Whole mitochondrial DNA sequences have been analyzed (not just HVRI and HVRII) from 53 humans with mtDNA from another primate species, chimpanzee, as an outgroup. This analysis showed important features which corresponded well with other studies on mtDNA and nuclear genes. The most recent common ancestor for human mtDNA was dated to 172 000 ± 50 000ya. There was separation between African and non-African mtDNA lineages and the most recent common ancestor for the non-African lineages was dated to c. 80 000 ya. The latter relatively recent date for the origin of non-African mtDNA lineages shows that H. sapiens migrated from Africa as much as 100 000 years after evolving there. Recently discovered hominid fossil evidence from Herto, Middle Awash, Ethiopia, has been interpreted as the immediate ancestors of anatomically modern humans and a new species name, H. sapiens idaltu, has been given to these fossils. H. sapiens idaltu has been dated to 160 000-154 000 ya and thus the gap between the genetic dating and the fossil evidence is being filled in. With the combination of the genetic and the new fossil evidence, there can be little doubt that modern humans evolved in Africa and multiregionalism is seen by many as a discredited hypothesis. The debate has moved on to other interesting questions concerning the direction and timing of the Out of Africa migration. Although the archaeological evidence for human migration is patchy or nonexistent in many regions of the Old World, the DNA evidence from present-day populations is again producing new insights into past events. All mtDNA haplogroups found in present-day populations are shown in a network analysis in Figure 2, with the geographical origin of each haplogroup shown in color. The Neanderthal mtDNA sequence is also included to show its relationship to modern human mtDNA, and the position of Mitochondrial Eve is also shown as belonging with the African mtDNA haplogroups. The L3* hap-logroup can be seen as giving rise to all non-African haplogroups (see below).
Which Way and When?
Extensive mtDNA research has been carried out in populations around the world to trace the route taken by H. sapiens out of Africa into the Old World (and indeed into colonization events in the New World and Oceania). Archaeological evidence has shown that modern humans were present in the Levant at c. 100-90 000 ya at the cave sites of Qafzeh and Skhul, but these are now seen as either an extension of African populations or a failed migration event. This northern route into Europe and Asia was not the one used by modern humans, and modern humans do not seem to have colonized Europe until much later, c. 45 000 ya. In fact, Australia seems to have been colonized before Europe (OSL and TL dates of 50 000-60 000ya have been obtained)!
All non-African mtDNA haplogroups coalesce to c. 80 000 ya - another way of saying this is that all non-African mtDNA haplogroups (those found in Asia, Europe, and Australia) are descended from an African ancestral mtDNA haplogroup. All African mtDNA haplotypes fall into three haplogroups. Using the mtDNA nomenclature, these are L1, L2, and L3, and these are further subdivided (e. g., L1a, L1b, etc). All non-African mtDNA haplogroups are derived from L3 - the origin of L3 itself is dated to 84 000 ya. Haplotypes derived from L3 in Asia are not found in the African L3 haplotypes (Figures 2 and 3). L3 in turn gave rise to two haplogroups called M and N approximately identical in age at c. 63 000 ya: each of these haplogroups is further subdivided into branches. The branches derived from M are called C, D, E, and
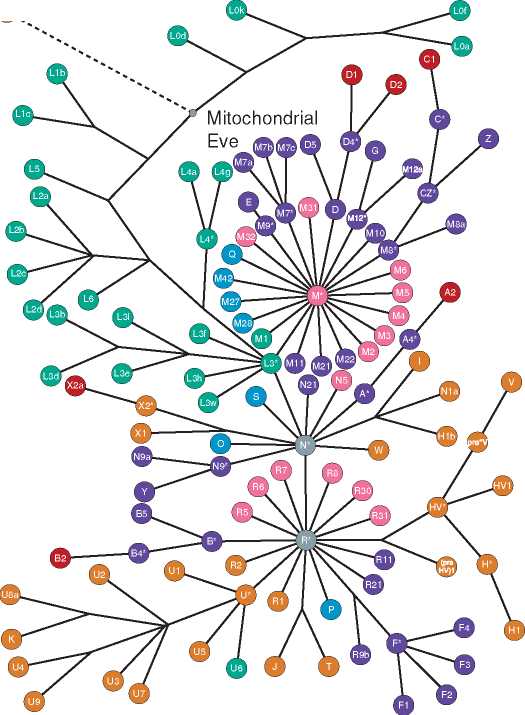
G, and these are found mainly in South and East Asia and America. The branches from N are A, B, F,
H, I, J, K, R, T, U, and V and these haplogroups are found mainly in European and West Asian populations (although A and B are also found in America, for example). Haplogroup R seems to have emerged rapidly within N at c. 60000ya. Eastern Africa has now been identified as the source population for the initial migration out of Africa, as Ethiopian populations have a high frequency of M haplotypes (variants of the M haplogroup) not found in sub-Saharan African populations. The genetic evidence also supports the theory of a single dispersal event out of Africa, rather than multiple ones.
How did humans manage to get to Australia before Europe? There is now archaeological evidence to support the mtDNA evidence that indicates a southern, ‘beachcombing’ route around the coast of Asia (Figure 4). The earliest evidence for the use of marine resources dates to 125 000 ya and is found at Abdur on the Eritrean coast of the Red Sea, so early modern humans were already able to exploit this subsistence strategy. In fact, a rough estimate has been made of the dispersal rate of humans using the southern route of 0.7-4 km yr-1, so that the time taken to reach SE Asia and Australia could have been quite rapid after the initial departure, perhaps only a couple of thousand years in all. Even the number of women
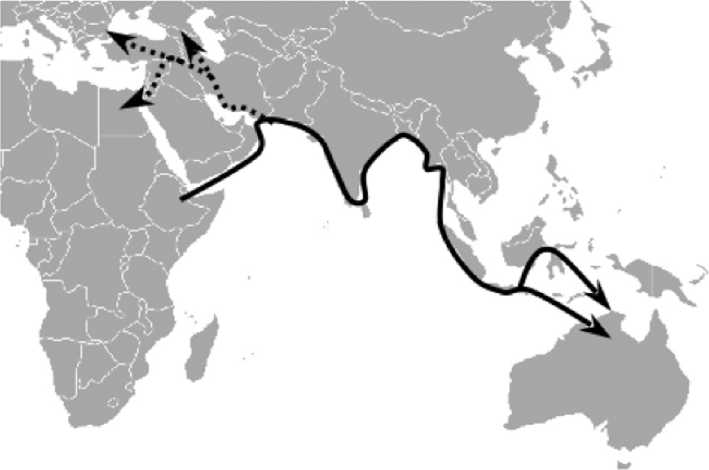
Figure 4 Map showing the southern beachcombing route Out of Africa, following the coastline to Australia (solid line), and the much later migration into Western Asia, Europe, and North Africa (dotted line).
Involved in the initial migration out of Africa has been estimated as about 600, but perhaps as low as 500 and as high as 2000. Although the true number may eventually be found to be higher or lower, this estimate gives us a feel for the size of the modern human population involved in this critical event in human prehistory. Various coastal populations considered by anthropologists to be ‘relicts’ of the Out of Africa migration have been sampled for mtDNA analysis, such as the Andaman Islanders, the Orang Asli, the Semang, the Senoi and Aboriginal Malays (all from Malaysia), the Papuans and Aboriginal Australians. If these populations are indeed descended from the initial Out of Africa dispersal that followed a coastal southern route, then their mtDNA haplogroups should coalesce to the same founder mtDNAs at 60 000 ya or earlier. If a northern route through the Levant was taken, then the coalescence time for their mtDNA haplogroups should be much more recent at 45 000-40 000 ya. The relict populations should also contain mtDNA haplotypes that are geographically distinct due to their isolation from later population movements.
Although a small sample number was used in this study, the results show that the Orang Asli and the Andamanese populations both contained M mtDNA haplotypes that branched off c. 60 000 ya, soon after the M haplogroup itself appeared in human populations. The Orang Asli also contained a number of M haplotypes that are virtually restricted to this population. Along with the analysis of the complete mtDNA genome from Papuans and Aboriginal
Australians, we now have strong genetic evidence for an early southern route of dispersal from Africa into the Old World. One genetic estimate for the time of arrival in Australia is 70 000ya. Again it is a case where the archaeological evidence needs to catch up with the genetics. A recent review by Mellars sets out the archaeological evidence known so far and suggests that this also supports the case for the southern route and a single dispersal event. The detailed archaeology of the coastal southern route and the identification of the earliest sites in Australia await discovery, although changes in sea level may mean that many sites may lie below the sea.
Into Europe - Eventually
The presence of modern humans in the Levant at 90 000-100 000 ya has been interpreted by palaeoanthropologists as either a failed attempt to colonize this region or as merely an extension of the African population. The earliest archaeological evidence for modern humans in Europe dates to c. 45 000 ya. The archaeology includes evidence of cognitive behavior that is associated with being ‘modern’, that is, the use of symbols, personal ornaments, art, formal burial and so forth. It has been claimed that the European evidence is the first to show a change in human behavior sometimes dubbed ‘the human revolution’. However, why should a species that had been around for perhaps 140 000 years wait until it had got to a very small continental cul de sac (for such is Europe) to suddenly show its modernity?
Archaeological evidence from Africa shows that certain behaviors (personal ornaments and symbolism) were already existing at least 100 000-80 000 ya (the former date for perforated shells found at Skhul), while formal burial and cave paintings are attested in Australia 60 000-32 000 ya. The major constraint on European colonization by modern humans was climate, not cognitive abilities.
As seen above, European mtDNA haplogroups are derived from the N haplogroup, which gave rise rapidly to the R haplogroup. Both of these haplogroups emerged soon after the Out of Africa dispersal, probably in South Asia, 63-60 000 ya. But all European mtDNA haplogroups are descended from N and R. These haplogroups are dated to c. 50 000 ya. From R the haplogroups B, F, H, V, T, J, U, and K are derived: haplogroups A, I, W, and X originate from N. Some of these haplogroups are not found in European populations and so can be omitted from this account (A, B, F, and R itself). All of the remaining haplogroups can be found in the Near East, with coalescence dates that are much earlier than for the same haplogroups in Europe. This shows that the sources for European colonization by modern humans were to be found in Western Asia (Figure 4), and ultimately these in turn derived from haplogroups that originate in South Asia. Some of the haplogroups in the Near East, such as J, T, U5, and I, date to 50 000-45 000 ya. Colonists from the Near East entered Europe, bringing what were to become European haplogroups with them. The earliest seems to be haplogroup U5, dating to c. 50 000ya according to the molecular clock. Other haplogroups arrived in Europe much later, as much as 15 000 years afterwards. The Upper Palaeolithic colonization of Europe by modern humans (also known as Cro-Magnons, after the French site where skeletal remains were first identified) has been linked by some geneticists with the appearance of the Aurignacian stone tool industry, and also a second, later wave of migration has been suggested as responsible for the introduction of the Gravettian stone tool industry. The link between stone tool industries and colonizations was first made in a study of Y chromosome haplogroups, which are difficult to date compared with mtDNA haplogroups, and must therefore be viewed with caution. However, mtDNA data would seem to support the Y chromosome evidence, as the next mtDNA haplogroup to enter Europe, haplogroup HV, gives a coalescence date of c. 33 000 ya, which coincides with the earliest appearance of the Gravettian in north east Europe. Although archaeologists are understandably wary of equating material culture with specific human groupings, either by population or ethnicity, in this case we have to consider the possibility that new stone tool industries and associated new cultural activities were indeed introduced by new peoples into Europe.

 World History
World History









